Research
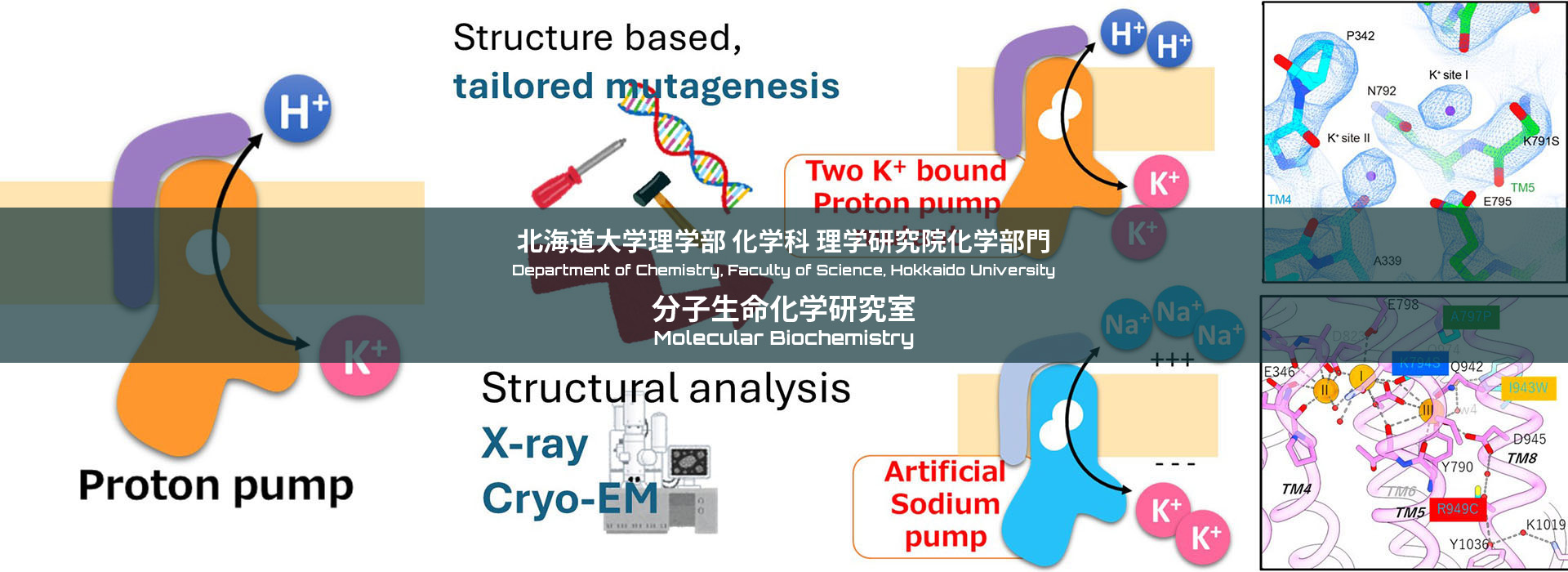
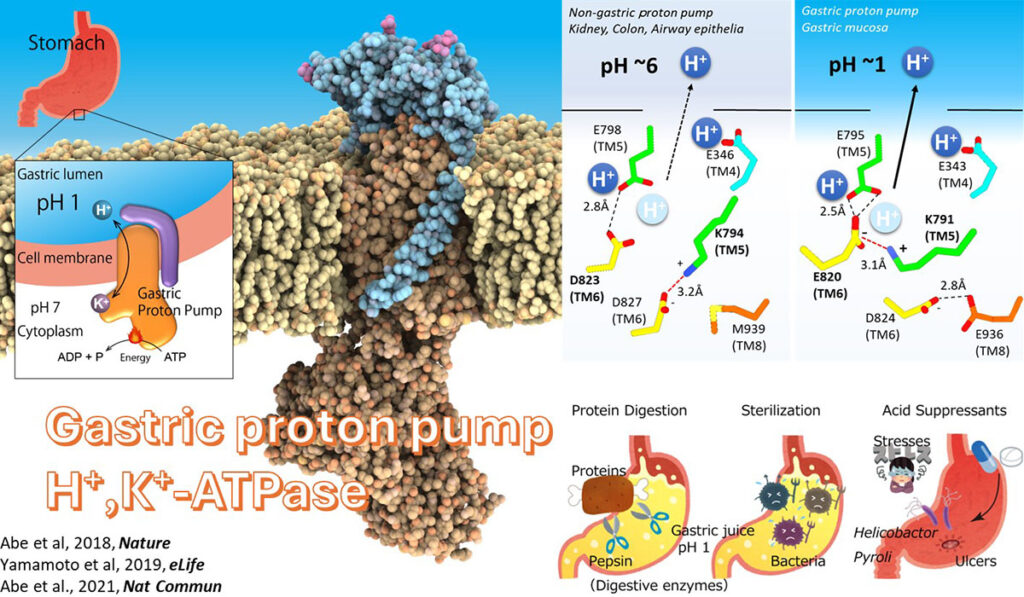
Gastric proton pump
When we have food intake, pH of our stomach reaches to 1. This highly acidic environment is indispensable for protein digestion, and also act as a barrier against various pathogen invasion via oral route. Conversely, too much acidification induces gastric ulcer. Gastric proton pump, a member of P-type ATPase family, is responsible for the gastric acid secretion, and therefore important drug target for the treatment of gastric acid-related diseases. Through structural and functional analysis, we will elucidate how it works at the level of chemistry.
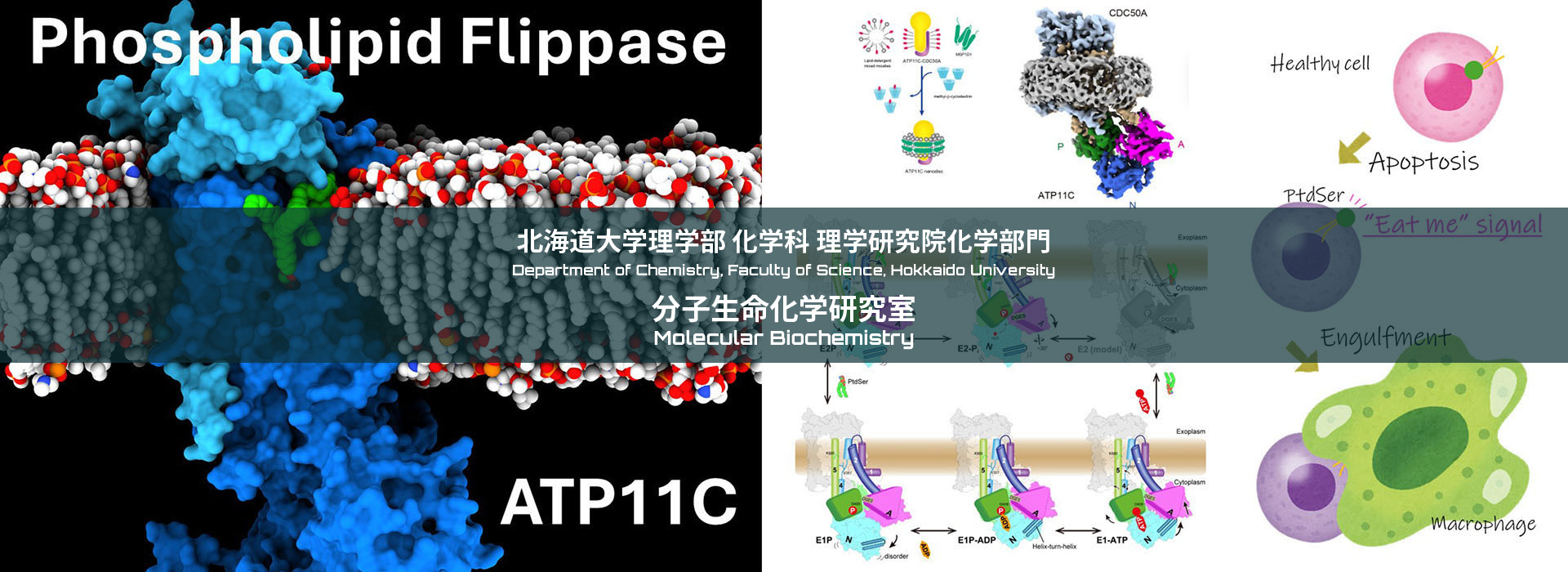
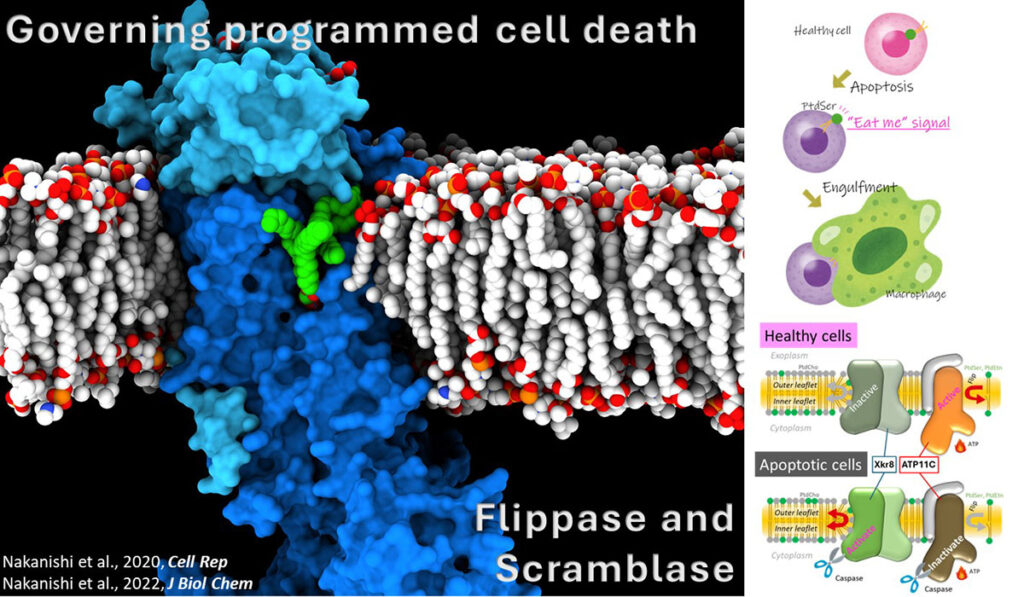
Phospholipid Flippase and Scramblase
In the plasma membrane of the living cell membrane, phosphatidylserine (PS) is confined in the cytoplasmic leaflet. When cell dies via apoptosis, PS is exposed to cell surface, which acts as “eat-me” signal of the cell, and dead cells are engulfed by macrophages without causing unnecessary inflammation. This system is maintained by concerted regulation of P4-ATPase flippases that translocate PS from outer leaflet to inner leaflet, and scramblases that mediate bidirectional translocation of PS. We attempt to reveal their molecular mechanism though structural and functional analysis.
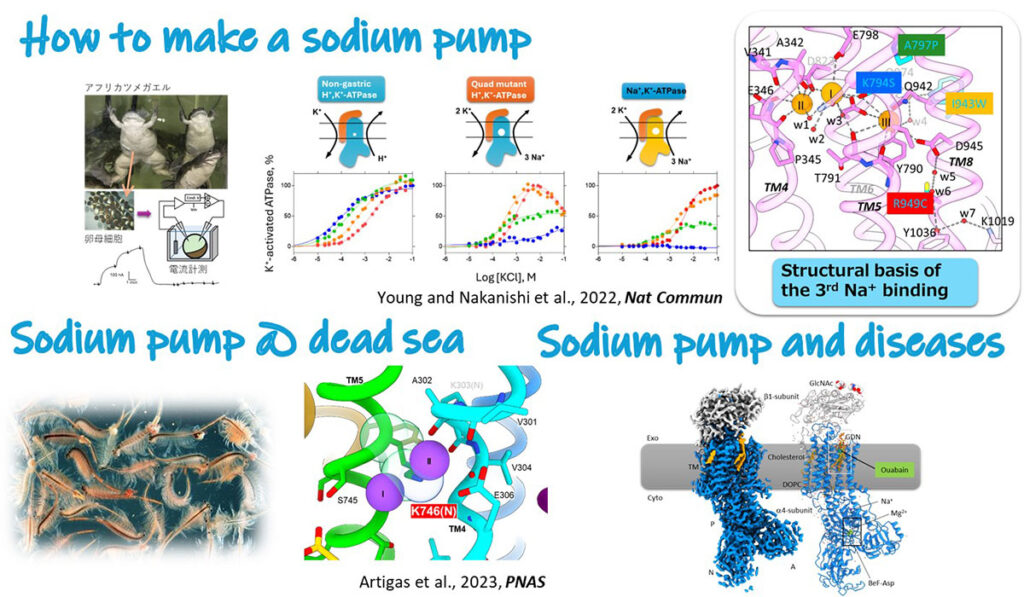
Sodium pump
In all animal cells, including ours human, concentrations of Na+ and K+ differs across the membrane. This electrochemical gradient of Na+/K+ is important basis for many physiological processes; providing a driving force for secondary transporters that allows cell to incorporate nutrients, regulation osmotic pressure, and being a source of the action potential in neuron. This is created by the membrane protein called sodium pump, Na+,K+-ATPase. Given the Na+/K+ gradient is involved in many physiological process, mutation on the sodium pump cause various diseases, including lapid-onset dystonia Parkinsonism and epilepsy. Understanding its molecular mechanism will lead to new therapeutic strategies.
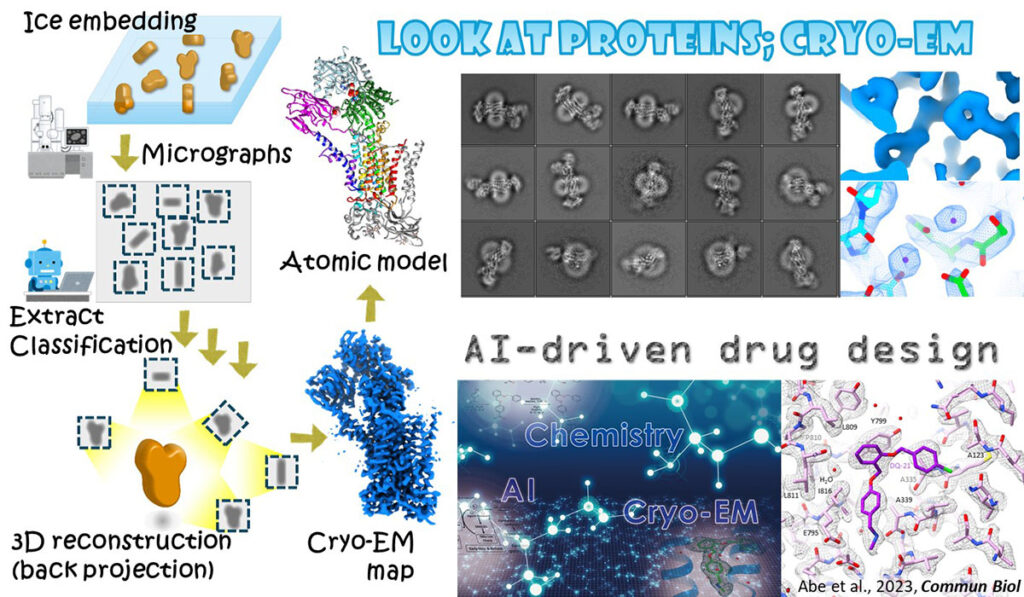
Cryo-EM
Proteins, and its substrates are far smaller than the wavelength of visible light, thus cannot be seen even using cutting-edge light microscope. However, electron beam does. Electron microscopy can visualize protein structure which provide indispensable information to elucidate their molecular mechanism at chemical level. Visualization of protein structure also allowed to design compounds that fit to the substrate binding site, that is, a potential drug. Employing generative AI technologies, we also attempt to develop a drug based on protein structure.